PHYSICIANS

A tissue test that improves patient stratification for repeat biopsy so you can focus on finding aggressive prostate cancer.
Confirm mdx Addresses Prostate Biopsy Sampling Error and the False-Negative Biopsy Dilemma
“Rule-in” high-risk men who have had a previous negative biopsy result, may be harboring undetected cancer (a false-negative biopsy result), and therefore may benefit from a repeat biopsy and appropriate treatment.
“Rule-out” otherwise cancer-free men from undergoing unnecessary repeat biopsies and screening procedures, helping to reduce complications, patient anxiety, and excessive healthcare expenses associated with these procedures.
Confirm mdx Key Benefits
Incorporating Confirm mdx into clinical practice improves patient stratification so you can focus on finding aggressive prostate cancer.
- Specific information for GS≥7 (GG≥2) prostate cancer
- Clinically validated in African American men
- Saves the healthcare system $588 per patient
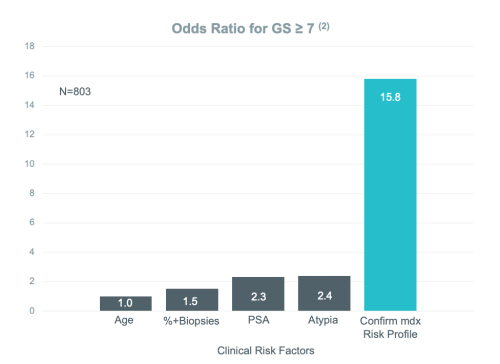
The Most Significant Predictor of Prostate Cancer Detection on Repeat Biopsy
Independently published clinical studies have shown that for men who have received a negative prostate biopsy result, Confirm mdx is the single most significant predictor of patient outcome among all currently available clinical factors, such as age, PSA level, and DRE results.1,2

- No additional office required
- Covered by Medicare
- Included in the NCCN Early Detection for Prostate Cancer Guidelines
Case Studies
Confirm mdx Improves Upon the Current Standard of Care
Under the current standard of care:
- Men with an elevated or rising PSA level (≥ 4.0 ng/ml) and/or abnormal digital rectal exam (DRE) are considered at high risk for cancer and often are referred for a prostate biopsy to determine if prostate cancer is present.
- The standard prostate biopsy procedure takes 10-12 core samples, and histopathological review by visual inspection under a microscope remains the gold standard for the diagnosis of prostate cancer.
- This schema, however, samples less than 1% of the entire prostate gland and results in limited histopathological analysis.
- Sampling error is an inherent and well-documented issue, with false-negative rates of prostate biopsy procedures reported as high as 25-35%.3,11

Of an estimated 500,000 biopsies performed each year, less than one-third actually result in a cancer finding, leaving more than 300,000 men with a negative biopsy reading but still facing elevated clinical risk factors.3,11,12 Concerns over inconclusive (false-negative) biopsy results, coupled with the high rate of clinically significant cancer detected upon repeat biopsy, pose a diagnostic dilemma:
- 43% of patients with negative histopathology on initial biopsy will undergo a repeat biopsy, many also continuing on to 3rd and 4th biopsies.
- Repeat biopsies are invasive procedures resulting in increased risk of infection and hospitalization.
- Significant costs are associated with unnecessary procedures and associated risks.13,15
For patients with a negative biopsy but with persistently elevated or rising PSA, abnormal DRE, or other risk factors, few options are currently available to guide a urologist in determining the patients need for an additional biopsy procedure is warranted. Fear of undetected prostate cancer leads to additional procedures, with many men being subjected to repeated invasive biopsies to rule-out the presence of cancer.
Fear of Undetected Cancer Leads to High Rate of Repeat Biopsy

Confirm mdx is a Well-Validated Epigenetic Test that Guides the Detection of Occult Prostate Cancer on a Patient’s Previously Biopsied Negative Tissue
- The use of Confirm mdx for prostate cancer detection using methylation-specific PCR (MSP) and cancer-associated epigenetic biomarkers to improve upon histopathology has been well validated in both scientific and clinical studies.
- DNA methylation, the most common and useful measure of epigenetic abnormality testing, is responsible for the silencing of key tumor suppressor genes. DNA methylation biomarkers associated with prostate cancer have been extensively evaluated. More than 55 studies on the Confirm mdx genes and technology have been published in peer-reviewed, scientific and medical journals.
- GSTP1 is the most intensely studied and widely reported epigenetic biomarker associated with prostate cancer diagnosis, encoding the glutathione S-transferase Pi 1 protein involved in detoxification, due to its high sensitivity and specificity.1,2,11
- Complementing GSTP1, methylation of the APC and RASSF1 genes is frequently found in prostate cancer, and these markers have demonstrated a “field effect” aiding in the identification of biopsies with false-negative histopathological results.1,2,11
- The concept of a field cancerization effect, when first reported in medical literature by Slaughter in 1953, described the changes in tissues surrounding cancer lesions and their association with development of tumors. Later, the term “field effect” evolved to include molecular changes in adjacent, benign-looking tissues. The epigenetic field effect is a molecular mechanism whereby cells adjacent to cancer foci can contain DNA methylation changes, which may be indistinguishable by histopathology, but detectable by MSP testing. The presence of epigenetic field effects associated with prostate cancer has been widely published and is the basis of activity for the Confirm mdx assay to aid in the detection of occult prostate cancer on previously biopsied, histopathologically negative tissue.15-22

Over 55+ Published Studies on the Test, Genes and Technology
Study of 211 African American Men
Evaluation of an Epigenetic Assay for Predicting Repeat Prostate Biopsy Outcome in African American Men96% Negative Predictive Value for High-grade Cancer
Risk Score Predicts High-Grade Prostate Cancer in DNA-Methylation Positive, Histopathologically Negative BiopsiesThe DOCUMENT Study: The Most Significant, Independent Predictor for Prostate Cancer in a Repeat Biopsy
Clinical Validation of an Epigenetic Assay to Predict Negative Histopathological Results in Repeat Prostate BiopsiesThe MATLOC Study Included 483 Men with an Initial Negative Biopsy, Followed by a Positive or Negative Repeat Biopsy within 30 Months
Clinical Utility of an Epigenetic Assay to Detect Occult Prostate Cancer in Histopathologically Negative Biopsies: Results of the MATLOC StudyReduced Rate of Repeated Prostate Biopsies Observed in ConfirmMDx
Reduced rate of Repeated Biopsies Observed in ConfirmMDx Clinical Utility Field StudyConfirmMDx Reduces Healthcare Costs and Helps to Differentiate Patients Who Should Undergo Repeat Biopsies
Budget Impact Model: Epigenetic Assay Can Help Avoid Unnecessary Repeated Prostate Biopsies and Reduce Healthcare Spending- Waterhouse RL Jr, Van Neste L, Moses KA, Barnswell C, Silberstein JL, Jalkut M, Tutrone R, Sylora J, Anglade R, Murdock M, Shiffman Z, Vandenberg T, Shah N, Carter M, Krispin M, Groskopf J, Van Criekinge W. Evaluation of an Epigenetic Assay for Predicting Repeat Prostate Biopsy Outcome in African American Men. Urology. 2018 Apr 13. pii: S0090-4295(18)30315-7. doi: 10.1016/j.urology.2018.04.001.
- Van Neste L, Groskopf J, Grizzle WE, Adams GW, DeGuenther MS, Kolettis PN, Bryant JE, Kearney GP, Kearney MC, Van Criekinge W, and Gaston SM. Epigenetic Risk Score Improves Prostate Cancer Risk Assessment. The Prostate. 2017 Sep;177(12):1259-64.
- Van Neste L, Partin AW, Stewart GD, Epstein JI, Harrison DJ, and Van Criekinge W. Risk Score Predicts High-Grade Prostate Cancer in DNA-Methylation Positive, Histopathologically Negative Biopsies. The Prostate. 2016 Sep;176(12):1078-87.
- Maldonado L, Brait M, Layo M, Sullenberger L, Wang K, Pekoe SB, Rosenbaum E, Howard R, Toubajo A, Albadine R, Netto GJ, Hoque MO, Platz EA, Sidrasky D. GSTP1 Promoter Methylation is Associated with Recurrence in Early Stage Prostate Cancer. The Joumal of Urology. 2014 Nov;192:1-7.
- Partin AW, Van Neste L, Klein EA, Marks LS, Gee JR, Troyer DA, Rieger-Christ K, Jones JS, Magi-Galluzzi C, Mangold LA, Track BJ, Lance RS, Bigley JW, Van Criekinge W, Epstein JI. Clinical Validation of an Epigenetic Assay to Predict Negative Histopathological Results in Repeat Prostate Biopsies. The Joumal of Urology. 2014 Oct;192(4):1081-7.
- Wojno KJ, Costa FJ, Cornell RJ, Small JD, Pasin E, Van Criekinge W, Bigley JW, Van Neste L. Reduced rate of Repeated Biopsies Observed in ConfirmMDx Clinical Utility Field Study. Am Health Drug Benefits. 2014 May;7(3):129-134.
- La Rosa FG, Jones C, Arangua P, Crawford ED, Van Neste L. Finding Occult Prostatic Cancer: The Value of Transperineal Mapping Biopsies and Epigenetic Assays. Joumal of Oncopathology. 2014 March;2.
- Crawford ED, Ventii K, Shore ND. New bio-markers in prostate cancer. Oncology (vVilliston Park). 2014 Feb;28(2):135-42.
- Van Neste L, Herman JG, Van Criekinge W. Improving Prostate Cancer Management: Clinical Lltility of Epigenetic for Diagnosis and Prognosis. The Joumal of Oncopathology. 2013 Apr;1 :29-32.
- Stewart GD, Van Neste L, Delvenne P, Delree P, Delga A, McNeil! SA, O’Donnell M, Clark J, Van Criekinge W, Bigley J, Harrison JD. Clinical Utility of an Epigenetic Assay to Detect Occult Prostate Cancer in Histopathologically Negative Biopsies: Results of the MATLOC Study. Joumal of Urology. 2013 Mar;189(3):1110-6.
- Aubry W, Lieberthal R, Willis A, Bagley G, Willis S, Layton A. Budget Impact Model: Epigenetic Assay Can Help Avoid Unnecessary Repeated Prostate Biopsies and Reduce Healthcare Spending. Am Health Drug Benefits. 2013 Jan-Feb;6(1):15-24.
- Van Neste L, Bigley J, Toll A, Otto G, Clark J, Delree P, Van Criekinge W, Epstein, JI. A tissue biopsy-based epigenetic multiplex PCR assay for prostate cancer detection. BMC Urol. 2012;12:16.
- Jeronimo C, Usadel H, Henrique R, Oliveira J, Lopes C, Nelson WG, Sidransky D. Quantitation of GSTP1 methylation in non-neoplastic prostatic tissue and organ-confined prostate adenocarcinoma. J Natl Cancer. 2001 Nov 21 ;93(22):1747-52.
- Trujillo KA, Jones AC, Griggith JK, Bisoffi M. Markers of Field Cancerization: Proposed Clinical Applications in Prostate Biopsies. Prostate Cancer. 2012;2012:302894.
- Van Neste L, Herman JG, Otto G, Bigley JW, Epstein JI, and Van Criekinge V. The Epigenetic promise for prostate cancer diagnosis. Prostate. 2012 Aug 1 ;72(11):1248-61.
- Track BJ, Brotzman MJ, Mangold LA, Bigley JW, Epstein JI, Mcleod D, Klein EA, Jones JS, Wang S, McAskill T, Mehorta J, Raghavan B, Partin AW. Evaluation of GSTP1 and APC methylation as indicators for repeat biopsy in a high-risk cohort of men with negative initial prostate biopsies. BJU. Int. 2012; Jul;110(1):56-62.
- Devaney J, Stirzaker C, Qu W, et al. Epigenetic deregulation across chromosome 2q14.2 differentiates normal from prostate cancer and provides a regional panel of novel DNA methylation cancer biomarkers. Cancer Epidemiol Biomarkers Prev. 2011 Jan;20(1 ): 148-59.
- Steiner I, Jung K, Schatz P, Horns T. Wittschieber D, Lein M, Dietel M, Erbersdobler A. Gene Promoter Methylation and Its Potential Relevance in Early Prostate Cancer Diagnosis. Pathobio/ogy. 2010;77(5): 260-6.
- Troyer DA, Lucia MS, de BrLiine AP, et al. Prostate Cancer Detected by Methylated Gene Markers in Histopathologically Cancer-Negative Tissues from Men with Subsequent Positive Biopsies. Cancer Epidemiol Biomarkers Prev. 2009 Oct; 18(10):2717 -22.
- Zon G, Barker MA, Kaur P, et al. Formamide as a denaturant for bisulfite conversion of genomic DNA: Bisulfite sequencing of the GSTPi and RARbeta2 genes of 43 formalin-fixed paraffin-embedded prostate cancer specimens. Anal. Biochem. 2009 Sep 15;392(2):117-25.
- Ahmed H, Cappello F, Rodolico V, Vasta GR. Evidence of Heavy Methylation in the Galectin 3 Promoter in Early Stages of Prostate Adenocarcinoma: Development and Validation of a Methylated Marker for Early Diagnosis of Prostate Cancer. Transl. Oneal. 2009 Aug 18;2(3):146-156.
- Mehrotra J, Varde S, Wang H, et al. Quantitative, spatial resolution of the epigenetic field effect in prostate cancer. Prostate. 2008 Feb 1 ;68(2):152-60.
- Periy AS, Loftus B, Moroose R, et al. In silica mining identifies IGFBP3 as a novel target of methylation in prostate cancer. Br J Cancer. 2007 May 21;96(10):1587-1594.
- Eilers T, Machtens S, Tezval H, et al. Prospective diagnostic efficiency of biopsy washing DNA GSTP1 island hypermethylation for detection of adenocarcinoma of the prostate. Prostate. 2007;67:757–63.
- Bastian PJ, Ellinger J, Heukamp LC, et al. Prognostic value of CpG island hypermethylation at PTGS2, RAR-beta, EDNRB, and other gene loci in patients undergoing radical prostatectomy. Eur. Urol. 2007 Mar;51(3):665-74.
- Cho NY, Kim BH, Choi M, et al. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol. 2007 Feb;211 (3):269-77.
- Vanaja DK, Ballman KV Morlan BW, et al. PDLIM4 repression by hypermethylation as a potential biomarker for prostate cancer. C/in. Cancer Res. 2006 Feb 15;12(4):1128-36.
- Yegnasubramanian S, Lin X, Haffner MC, et al. Combination of methylated-DNA precipitation and methylation-sensitive restriction enzymes (COMPARE-MS) for the rapid, sensitive and quantitative detection of DNA methylation. Nucleic Acids Res. 2006 Feb 9;34(3):e19.
- Henrique R, Jer6nimo C, Teixeira MR, et al. Epigenetic Heterogeneity of High-Grade Prostatic Prostate Carcinogenesis lntraepithelial Neoplasia: Clues for Clonal Progression in Prostate Carcinogenesis. Mo/. Cancer Res. 2006 Jan;4(1):1-8.
- Enokida H, Shiina H, Urakami S, et al. Multigene methylation analysis for detection and staging of prostate cancer. Clin. Cancer Res. 2005 Sep 15;11 (18):6582-8.
- Bastian PJ, Palapattu GS, Lin X, et al. Preoperative Serum DNA GSTP1 CpG Island Hypermethylation and the Risk of Early Prostate-Specific Antigen Recurrence Following Radical Prostatectomy. C/in. Cancer Res. 2005 Jun 1 ;11 (11):4037-43.
- Jeronimo C, Varzim G, Henrique R, et al. 1105V Polymorphism and Promoter Methylation of the GSTP1 Gene in Prostate Adenocarcinoma. Cancer Epidemiol Biomarker Prev. 2005 May;11(5):445-50.
- Jeronimo C, Henrique R, Hoque MO, et al. A Quantitative Promoter Methylation Profile of Prostate Cancer. Clin. Cancer Res. 2004 Dec 15;10(24):8472-8.
- Bernardini S, Miano R, lori R, et al. Hypermethylation of the CpG islands in the promoter region of the GST.P1 gene in prostate cancer: a useful diagnostic and prognostic marker? Clin. Chim. Acta. 2004 Dec;350(1-2):181-8.
- Flori AR, Steinhoff C, Muller M, et al. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br J Cancer 2004: Aug 31 ;91 (5):985-94.
- Tokumaru Y, Harden SV, Sun DI, et al. Optimal Use of a Panel of Methylation Markers with GSTP1 Hypermethylation in the Diagnosis of Prostate Adenocarcinoma. Clin. Cancer Res. 2004 Aug 15; 10(16): 5518-22.
- Zhou M, Tokumaru Y, Sidransky D, Epstein JI. Quantitative GSTP1 Methylation Levels Correlate with Gleason Grade and Tumor Volume in Prostate Needle Biopsies. J Ural. 2004 Jun;171 (6 Pt 1 ):2195-8.
- Woodson K, Hanson J, Tangrea J: A survey of gene-specific methylation in human prostate cancer among black and white men. Cancer Lett. 2004 Mar 18;205(2):181-8.
- Yegnasubramanian S, Kowalski J, Gonzalgo ML, et al. Hypermethylation of CpG Islands in Primary and Metastatic Human Prostate Cancer. Cancer Res. 2004 Mar 15;64(6):1975-86.
- Kang GH, Lee S, Lee HJ, et al. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. J Pathol. 2004 Feb;202(2):233-40.
- Harden SV, Sanderson H, Goodman SN, et al. Quantitative GST-P1 methylation and the detection of prostate adenocarcinoma in sextant biopsies. J. Natl. Cancer Inst. 2003 Nov 5;95(21):1634-7.
- Nakayama M, Bennett CJ, Hicks JL, et al. Hypermethylation of the human glutathione S-transferase-pi gene (GST-P1) CpG island is present in a subset of proliferative inflammatory atrophy lesions but not in normal or hyperplastic epithelium of the prostate: a detailed study using laser-capture microdissection. Am. J. Pathol. 2003 Sept;163(3):923-33.
- Yamanaka M, Watanabe M, Yamada Y, et al. Altered Methylation of Multiple Genes in Carcinogenesis of The Prostate. Int. J. Cancer. 2003 Sept 1 ;106(3):382-7.
- Woodson K, Hayes R, Wideroff L, et al. Hypermethylation of GST-P1, CD44, and E-cadherin genes in prostate cancer among US Blacks and Whites. Prostate. 2003 May 15;55(3):199-205.
- Harden SV, Guo Z, Epstein JI, et al. Quantitative GST-P1 methylation clearly distinguishes benign prostatic tissue and limited prostate adenocarcinoma. J. Urol. 2003 Mar;169(3):1138-42.
- Konishi N, Nakamura M, Kishi M, et al. DNA hypermethylation status of multiple genes in prostate adenocarcinomas. Jpn. J. Cancer Res. 2002 Jul;93(7):767-73.
- Chu DC, Chuang CK, Fu JB, et al. The use of real-time quantitative polymerase chain reaction to detect hypermethylation of the CpG islands in the promoter region flanking the GST-Pl gene to diagnose prostate carcinoma. J. Urol. 2002 Apr;167(4):1854-8.
- Goessl C, Muller M, Heicappell R, et al.: Methylation-specific PCR for detection of neoplastic DNA in biopsy washings. J Pathol. 2002 Mar;196(3):331-4.
- Maruyama R, Toyooka S, Toyooka KO, et al. Aberrant Promoter Methylation Profile of Prostate Cancers and Its Relationship to Clinicopathological Features. Clin. Cancer Res. 2002 Feb;8(2):514-9.
- Un X, Tascilar M, Lee WH, et al. GSTP1 CpG Island Hypermethylation Is Responsible for the Absence of GSTP1 Expression in Human Prostate Cancer Cells. Am. J. Pathol. 2001 Nov; 159(5):1815-26.
- Jeronimo C, Usadel H, Henrique R, et al. Quantitation of GST-P1 methylation in non-neoplastic prostatic tissue and organ-confined prostate adenocarcinoma. J. Natl. Cancer. 2001 Nov 21; 93(22):1747-52.
- Goessl C, Krause H, Muller M, et al. Fluorescent methylation-specific polymerase chain reaction for DNA-based detection of prostate cancer in bodily fluids. Cancer Res. 2000 Nov 1; (60)21 :5941-5.
- Santourlidis S, Florl A, Ackermann R, et al. High frequency of alterations in DNA methylation in adenocarcinoma of the prostate. Prostate. 1999 May 15;39(3):166-7 4.
- Brooks JD, Weinstein M, Lin X, et al. CG island methylation changes near the GST-P7 gene in prostatic intraepithelial neoplasia. CancerEpidemiol Biomarkers Prev. 1998 Jun;7(6):531-6.
- Lee WH, Isaacs WB, Bova GS, et al. CG island methylation changes near the GST-P7 gene in prostatic carcinoma cells detected using the polymerase chain reaction: a new prostate cancer biomarker. Cancer Epidemiol Biomarkers Prev. 1997 Jun;6(6):443-50.
- Lee WH, Morton RA, Epstein JI, et al. Cytidine methylation of regulatory sequences near the n-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc. Natl. Acad. Sci. USA. 1994 Nov 22;91(24):11733-7.
Posters & Abstracts
- Van Neste L, Partin AW, Stewart GD, Epstein JI, Harrison DJ, and Van Criekinge W. Risk Score Predicts High-Grade Prostate Cancer in DNA-Methylation Positive, Histopathologically Negative Biopsies. The Prostate. 2016 Sep;176(12):1078-87.
- Partin AW, Van Neste L, Klein EA, Marks LS, Gee JR, Troyer DA, Rieger-Christ K, Jones JS, Magi-Galluzzi C, Mangold LA, Track BJ, Lance RS, Bigley JW, Van Criekinge W, Epstein JI. Clinical Validation of an Epigenetic Assay to Predict Negative Histopathological Results in Repeat Prostate Biopsies. The Joumal of Urology. 2014 Oct;192(4):1081-7.
- Merril J. Prostate cancer market snapshot: more than provenge. The Pink Sheet. November 22, 2010. Elsevier Business Intelligence Publications and Products. https://www.pharmamedtechbi.com/publications/the-pink-sheet/72/47/prostate-cancer-market-snapshot-more-then-provenge. Accessed January 29, 2013.
- Carroll P, Coley C, McLeod D, et al. Prostate-specific antigen best practice policy–part I: early detection and diagnosis of prostate cancer. Urology 2001; 57:217.
- Brawer MK, Chetner MP, Beatie J, et al. Screening for prostatic carcinoma with prostate specific antigen. J Urol 1992; 147:841.
- Catalona WJ, Smith DS, Ratliff TL, Basler JW. Detection of organ-confined prostate cancer is increased through prostate-specific antigen-based screening. JAMA 1993; 270:948.
- Crawford ED, DeAntoni EP, Etzioni R, et al. Serum prostate-specific antigen and digital rectal examination for early detection of prostate cancer in a national community-based program. The Prostate Cancer Education Council. Urology 1996; 47:863.
- Taneja et al.: The American Urological Association (AUA) Optimal Techniques of Prostate Biopsy and Specimen Handling. 2013.
- Shen et al.: Three-Dimensional Sonography With Needle Tracking – Role in Diagnosis and Treatment of Prostate Cancer. J. Ultrasound Med. 2008; Jun; 27(6): 895-905.
- Rabets JC et al.: Prostate cancer detection with office based saturation biopsy in a repeat biopsy population. The Journal of Urology. 2004; Jul; 172(1): 94–97.
- Kronz JD et al.: Predicting cancer following a diagnosis of high-¬‐grade prostatic intraepithelial neoplasia on needle biopsy: data on men with more than one follow-¬‐up biopsy. American Journal of Surgical Pathology 2001; Aug 25(8): 1079–1085.
- Based on MDxHealth management estimates taken from publicly available Medicare and other data.
- Trock BJ et al.: Evaluation of GSTP1 and APC Methylation as Indicators for Repeat Biopsy in a High-risk Cohort of Men with Negative Initial Prostate Biopsies. British Journal of Urology International 2012; July 110(1): 56-62.
- National Cancer Institute. Surveillance, Epidemiology and End Results. 2014 Prostate Cancer Statistics http://seer.cancer.gov/statfacts/html/prost.html.
- Pinsky PF, Crawford ED, Kramer BS, et al. Repeat prostate biopsy in the prostate, lung, colorectal and ovarian cancer screening trial. BJU Int. 2007;4:775-779.
- Mosquera J-M, Mehra R, Regan MM, et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res. 2009;15:4706-4711.
- Slaughter DP et al.: Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer 1953; Sep; 6(5): 963–8.
- Braakhuis BJ et al.: A Genetic Explanation of Slaughter’s Concept of Field Cancerization: Evidence and Clinical Implications. Cancer Res. 2003. Apr 15; 63(8): 1727-30.
- Van Neste L et al.: The Epigenetic Promise for Prostate Cancer Diagnosis. The Prostate 2011; 1; 72(11): 1248-61.
- Trujillo KA et al.: Markers of Field Cancerization: Proposed Clinical Applications in Prostate Biopsies. Prostate Cancer 2012; 2012:302894.
- Henrique R et al.: Epigenetic Heterogeneity of High-Grade Prostatic Prostate Carcinogenesis Intraepithelial Neoplasia: Clues for Clonal Progression in Prostate Carcinogenesis. Mol. Cancer Res. 2006; Jan; 4(1): 1-8.
- Zhou M et al.: Quantitative GSTP1 Methylation Levels Correlate with Gleason Grade and Tumor Volume in Prostate Needle Biopsies. J. Urol. 2004; Jun; 171 (6PT 1): 2195-8.
Contact a Local Representative
Questions about our tests or ready to set up a new account?
How To Order Confirm mdx
Ordering Confirm mdx for Prostate Cancer is simple and easy.
Report Access
Secure mdx is a safe and secure HIPAA compliant tool that gives you access to patient reports online.